Assays performed on day 13 Megakaryocytes. Count cells and resuspend in SFEM without growth factors at 1×10^6/mL . All assays should be done in comparison to a non-targeting crRNA control and if desired a targeting control (i.e. B2M)
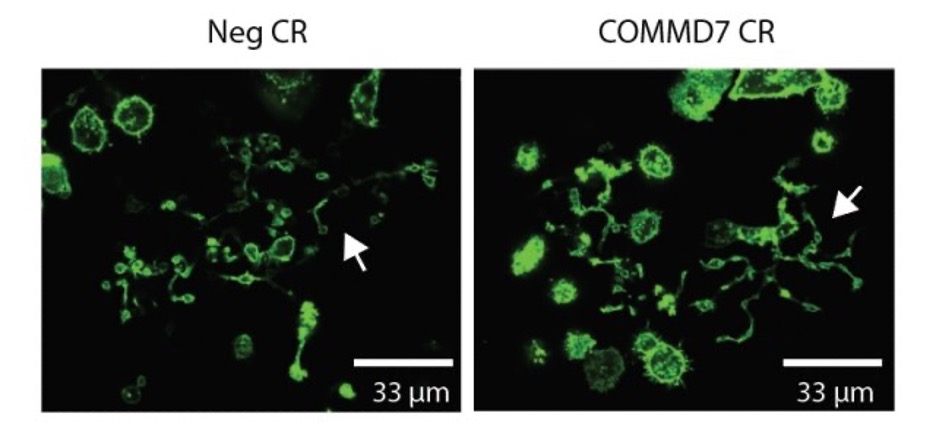
Proplatelet formation
- coat 8 well borosilicate chamber slide with 100 ug/mL fibrinogen diluted in PBS for 30 min at 37c
- remove fibrinogen, block with 2.5% human serum albumin for 20 minutes
- wash 3x PBS
- add 2.5×10^5 cells in 250 uL SFEM-T per well
- incubate overnight
- add 100 uL 4% PFA for 20 minutes at RT
- Fix with 2% paraformaldehyde
- wash 3x with PBS
- permeabilize cells with 0.1% triton x 100 for 5 minutes
- Wash 3x with PBS
- add phalloidin 488 and nuclear stain for 15 minutes at RT
- wash 3x with PBS, keep in PBS for imaging
- While blinded to treatment, image cells on fluorescent microscope, take ratio of # proplatelet forming vs total cells. Examples of proplatelet forming megakaryocytes are marked with white arrows in the images below.
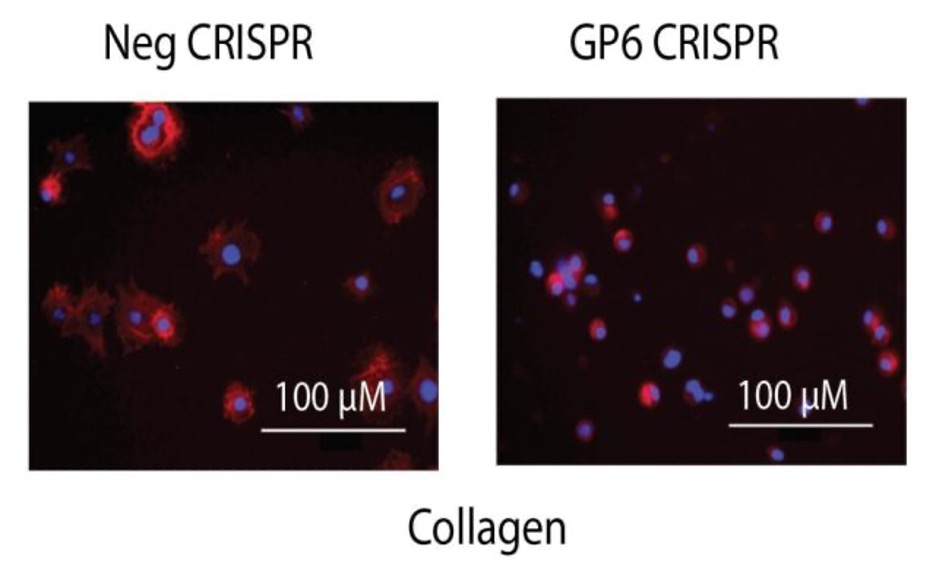
Spreading
- coat duplicate wells of 96 well plates with collagen (coated the night before) or fibrinogen
- Dilute collagen 1:10 in HBSS. 40 uL to each well, place at 4c overnight
- Thaw reconstituted fibrinogen at 37c. Dilute 1:10,000 in HBSS, 40 uL each well place for 30 minutes at 37c
- Wash wells 3 times with PBS
- Add 2.5×10^4 megakaryocytes to wells in 100 uL media.
- Incubate 1 hour at 37c
- gently remove unbound cells
- wash gently with PBS
- Fix with 2% paraformaldehyde
- process cells for imaging as in steps 8-12 of proplatelet assay
- While blinded to treatment, image cells on fluorescent microscope, take ratio of # spread vs round cells. In the example below most of the negative CRISPR treated cells are spread (egg on a pan) on colagen, whereas most GP6 KO cells are round.
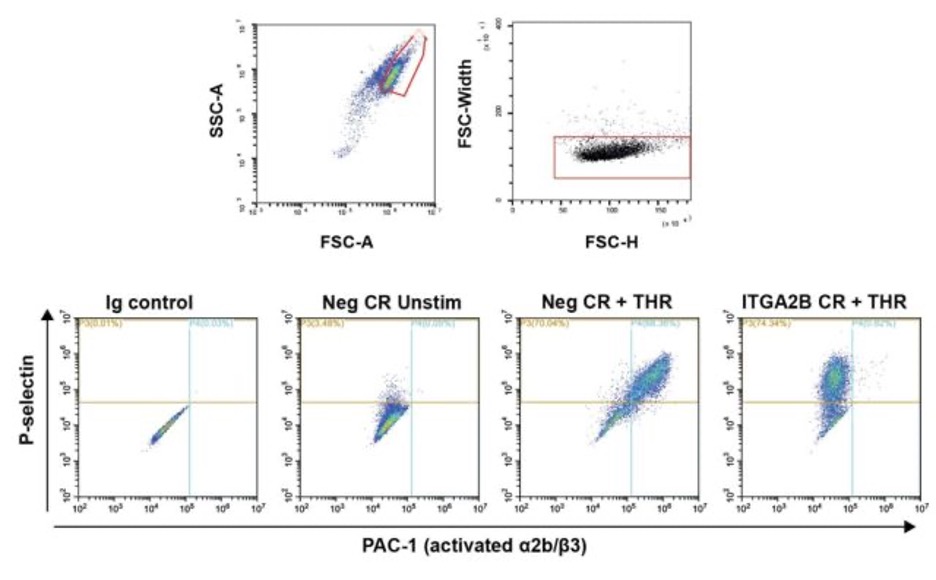
Flow cytometry phenotyping and assays of activation
We do activation assays in a round bottom polypropylene 96 well plate. Include an unstimulated activated and unactivated matching isotype control well for at least one of the conditions.
- Prealiquot 50 uL of phenotyping, activation, or fibrinogen binding antibody mix into individual wells. Use SFEM without cytokines:
Phenotyping mix
- 1 uL CD41-APC
- 2 uL CD42a-PE
- 1 uL CD61-FITC
- 46 uL SFEM
Activation mix
- 6 uL cd62-PE (granule marker)
- 10 uL PAC1-FITC (integrin activation)
- 5 uL CD63-V450 (dense granules/lysosomes, optional)
- 1 uL CD41-APC (MK marker)
- 28 uL SFEM
Fibrinogen binding mix
- 2 uL CD42a-PE
- 1 uL Fibrinogen-647
- 47 uL SFEM
2. Add vehicle (for unstimulated) or agonist, such as one of the following, to activation wells:
- 1 U/mL thrombin*
- 1.25 ug/mL convulxin
- 2 ug/mL CRP
- 200 nM 2-MesSADP
*Thrombin can’t be used in the fibrinogen binding assay without adding inhibitors to prevent clot formation
3. Immediately add 50 uL of cells (5×10^5) and pipet to mix (for many conditions use a multichannel pipetor
4. Incubate at 37c for 15 minutes
5. Fix with 100 uL 4% PFA (2% final)
6. Analyze by flow cytometry
Gate on higher FSC, lower SSC population as described by https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6886406/ . Further gate on singlets using FSC-H and FSC-W. Set phenotype marker (CD41, CD61,CD42a) or activation gates (PAC-1, P-selectin, or CD63) based on isotype controls. Record % positive cells and gMFI of the gated cells. Gating examples shown below for negative control vs ITGA2B KO cells.